Started in 1990, The U.S. Human Genome Project has had a major impact on the development of technologies for mass DNA sequencing. Initially based in the United States, the Human Genome Project soon expanded to involve many other countries. Not only did it invest heavily in the development of better DNA sequencing technologies, but it also led to the development of new DNA sequencing strategies for sequencing entire genomes.
As a result of the Human Genome Project, the two
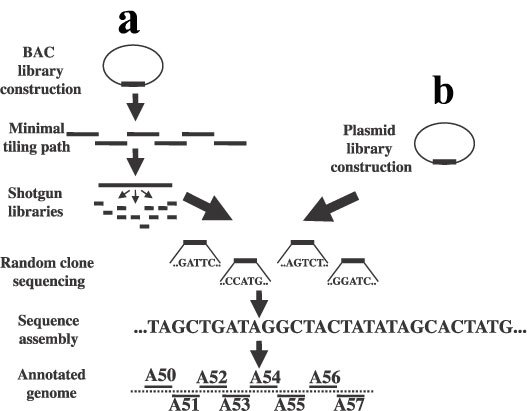
main basic strategies of sequencing DNA on a large scale, map-based sequencing and whole-genome shotgun sequencing, have been developed in the race to sequence the human genome. In map-based
sequencing, a physical map of the genes is made with a technique called
fluorescence in situ hybridization (FISH) and genetic markers are subsequently mapped. Several different copies of the DNA to be sequenced are
then cut up with several different restriction enzymes and sequenced. Fragments can be sequenced based on their overlap and the original genome
can be recreated, with the physical and genetic maps serving as place markers. This was
the main strategy used by the Human Genome Project. Whole-genome shotgun sequencing, a strategy promoted by the
company Celera Genomics, skips the steps
of constructing physical and genetic maps. Instead, it relies on the computational
ability of powerful computers to reconstruct the original genome. (8)
Although
Celera Genomics and the Human Genome Project each argue the advantages of their
approaches, in the end, both probably benefited from the work of the other.
(The end result was a tie between the two efforts because the draft genome sequences of both projects were published in the same week in the major science journals Science and Nature.) (21) (11)
As shown, shotgun sequencing (b) does not require many prepatory steps, as compared to the map-based sequencing (a), instead using powerful algorithms to later stitch together the genome. |
The Human Genome Project has also led to the establishment of major databases of the DNA sequences of more than 100,000 different organisms for worldwide researchers and also accessible to the general public. Worldwide laboratories submit their data to GenBank, a collaborative database involving areas all over the world including Japan, Europe, and the U.S. The raw DNA sequence data in GenBank has been growing at an exponential rate. Researchers can then search GenBank for the annotated gene sequences of various organisms. For example, click here to view raw data of the human chromosome 14, or view a graphical representation here. The main page of GenBank is here. (33)