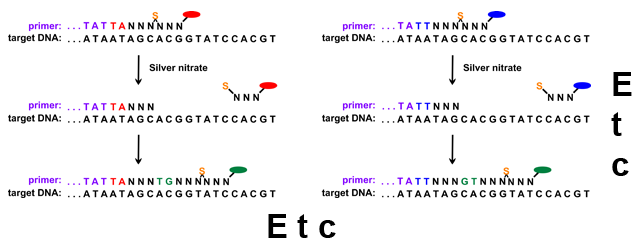
SOLiD DNA sequencing is a very complicated process, so just remember it uses an enzyme called ligase. Ligase, unlike DNA polymerase, does not add individual bases. Instead, it adds a whole segment of bases that is complementary to the DNA being sequenced. Also, remember that it has a built-in proofreading capability which gives it very low error rates. We'll gloss over some of the finer details.
This diagram illustrates steps 3 and 4, in which ligase adds a short complementary DNA fragment in the left column, and then shifts one base over in the right column.
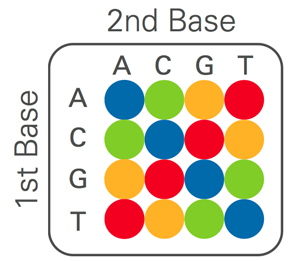
This diagram shows the assignment of the four colors to each two-base combination. |
|
Now, we have data of four different colors. To make
things more complicated, although you would logically think each color should
correspond to one base, each color actually corresponds to TWO bases, the two
bases at the beginning of each DNA fragment, even though there are only FOUR
different colors. If you do the math, you will find that there really should be
SIXTEEN different colors to correspond to every possible two-base combination
(4 times 4, since there are 4 possible bases per position). However, this
problem is solved by assigning four different two-base combinations to each of
the four different colors. Since we had five different reading frames, by doing
some complex analysis, we are able to determine the sequence from this data,
while at the same time double-checking the sequencing several times. (47)