This unique method involves the formation of
DNA "nanoballs", before using the enzyme ligase to sequence the DNA as in SOLiD sequencing.
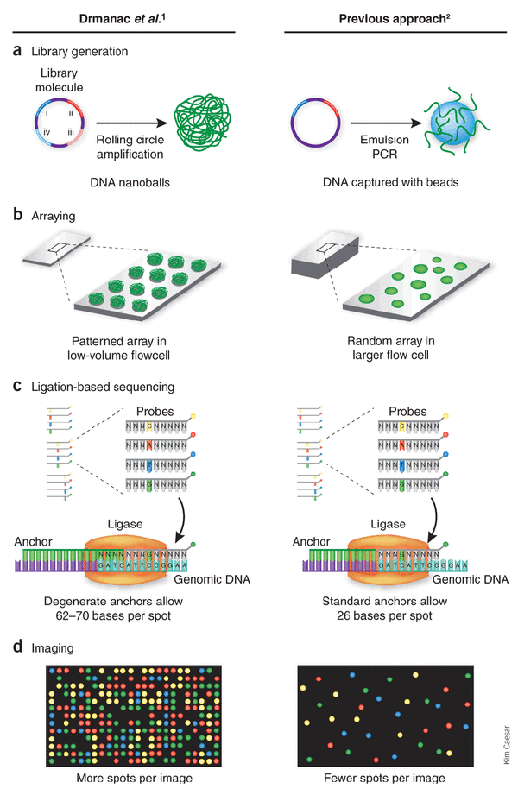
Above, the diagram shows the main advantage of DNA nanoballs versus other methods which use emulsion PCR: much greater amplification and color intensity.
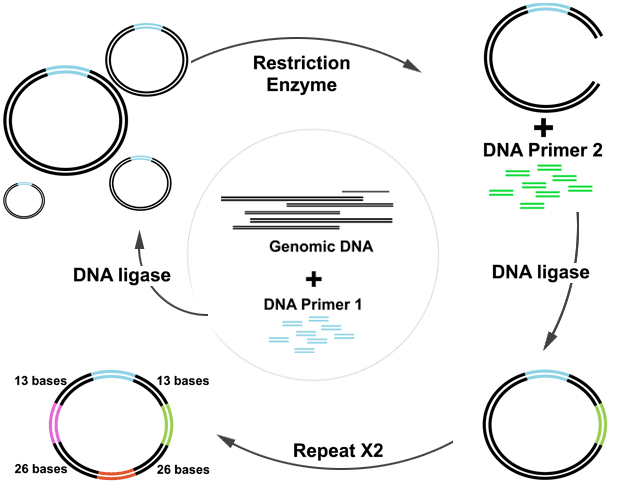
Above, an illustration of the addition of the four primers to form loops of DNA in step 1.
- DNA ligase first joins short, linear DNA fragments together at both ends with another shorter DNA fragment of known sequence called aprimer to form closed loops of DNA. Special restriction enzymes cut each loop at a specific area exactly 13 bases in both directions, with DNA ligase inserting in two more primers, one at each cut. Finally, a second enzyme adds one more primer, also at a known location, bringing the total number of inserted primers to 4.
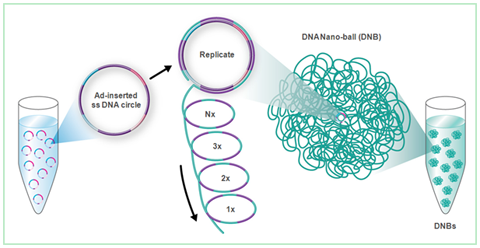
At left, the amplification process described in step 2 is illustrated in detail. The ssDNA circles are the prepared DNA fragments forming closed loops. |
2. A
special DNA polymerase replicates the looped DNA, and when it finishes one
circle, it doesn't stop- it continues replication by peeling off its
previously copied DNA. This copying process continues over and over,
forming a DNA nanoball, a large mass of the repeating DNA to be
sequenced all connected together.
3. Like SOLiD, Complete Genomics also uses ligase for sequencing- it also requires putting in all five-base possible DNA fragments (over 1000!). At each DNA-copy on the DNA nanoball, a ligase puts in the correct five-base DNA fragment complementary to the DNA to be sequenced. At the end of the five-base DNA fragment is a dye.
4. There are four different colored dyes, with each labeling a different nucleotide. In the first cycle, the dye represents the first nucleotide on the five-base DNA fragment.
5. The color of the dye is recorded and the entire five-base DNA fragment is removed. In the next cycle, all possible five-base DNA fragments are again inserted, except this time the dye at the end represents the second nucleotide on the five-base DNA fragment. After this cycle is performed five times, five bases will have successfully been sequenced.
6. This process can be performed several times on each DNA nanoball for a total of 66 different bases per nanoball. (34) (46)
3. Like SOLiD, Complete Genomics also uses ligase for sequencing- it also requires putting in all five-base possible DNA fragments (over 1000!). At each DNA-copy on the DNA nanoball, a ligase puts in the correct five-base DNA fragment complementary to the DNA to be sequenced. At the end of the five-base DNA fragment is a dye.
4. There are four different colored dyes, with each labeling a different nucleotide. In the first cycle, the dye represents the first nucleotide on the five-base DNA fragment.
5. The color of the dye is recorded and the entire five-base DNA fragment is removed. In the next cycle, all possible five-base DNA fragments are again inserted, except this time the dye at the end represents the second nucleotide on the five-base DNA fragment. After this cycle is performed five times, five bases will have successfully been sequenced.
6. This process can be performed several times on each DNA nanoball for a total of 66 different bases per nanoball. (34) (46)